BREAKING: A groundbreaking study from KU Leuven has just revealed a revolutionary deep learning model that transforms how scientists analyze the 3D microstructure of fruit tissues. Published in Plant Phenomics on July 5, 2025, this innovative approach significantly outperforms traditional 2D methods, offering a new, automated way to investigate vital plant processes.
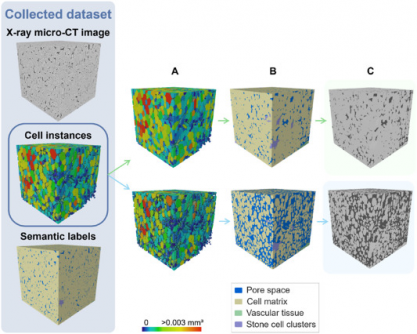
The study, led by Pieter Verboven, addresses longstanding challenges in plant tissue analysis. Traditional microscopy requires extensive preparation and limits the scope of observation, hindering research on crucial metabolic processes. However, the newly developed 3D model utilizes X-ray micro-CT technology to achieve non-destructive imaging, providing a clearer and more comprehensive view of fruit microstructures.
The urgency of this research cannot be overstated. The model has achieved remarkable accuracy, with an Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears, surpassing previous benchmarks and demonstrating its potential to revolutionize plant biology. This development allows for precise and automated segmentation of plant tissues, crucial for understanding how cellular arrangements affect fruit quality, storability, and susceptibility to disorders such as browning.
Utilizing a state-of-the-art 3D panoptic segmentation framework based on the 3D extension of Cellpose and a 3D Residual U-Net, the model performs dual segmentation tasks. It accurately predicts gradient fields to separate individual parenchyma cells while classifying various tissue types such as cell matrix, pore space, and vascular structures. The research team employed extensive datasets augmented with synthetic morphological variations, validating the model’s effectiveness against conventional 2D methods.
Visual validation confirmed the model’s capability to accurately detect vascular bundles in varieties like ‘Kizuri’ and ‘Braeburn’ apples, while also achieving smooth segmentation of stone cell clusters in ‘Celina’ and ‘Fred’ pears. This level of precision is a game changer for plant scientists, providing a powerful tool for exploring how microscopic structures influence crucial factors such as nutrient transport and stress responses.
As the scientific community responds to this innovation, the implications extend beyond fruit research. The technology presents a scalable framework for studying tissue development and ripening across various crops, making it an accessible solution for integrating artificial intelligence into agricultural and food science research.
Looking ahead, the research team anticipates further developments in data augmentation techniques to enhance model performance. However, they acknowledge existing challenges related to dataset imbalance and domain shifts that may require additional focused efforts.
This pioneering work is not only positioned to accelerate research but also to reduce manual labor involved in tissue characterization, making it a significant leap forward in the intersection of technology and plant science. The urgency of this research highlights its essential role in addressing key scientific challenges in agriculture and food production.
Stay tuned for more updates as the implications of this study unfold, potentially reshaping how we understand plant physiology and its applications in food science.