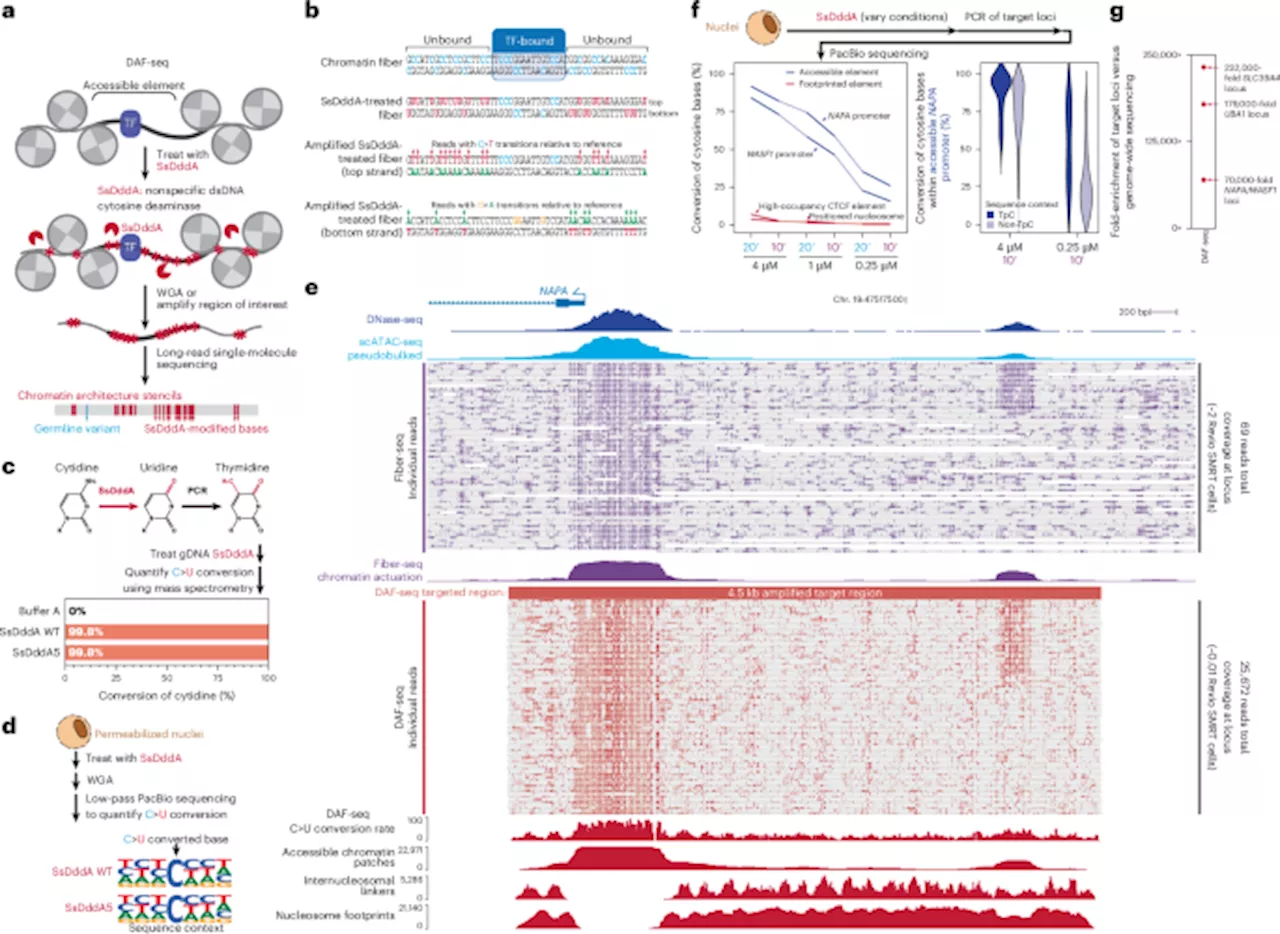
Scientists at the University of Washington have developed a groundbreaking technique known as Deaminase-Assisted single-molecule chromatin Fiber sequencing, or DAF-seq. This innovative method enables researchers to map single-cell chromatin fiber architectures with remarkable precision, offering insights into gene regulation at an unprecedented resolution.
DAF-seq addresses a significant challenge in understanding gene regulation, which is influenced by the co-binding of proteins along chromatin fibers within single cells. Until now, the variability of this occupancy between haplotypes and individual cells in diploid organisms remained poorly understood. With DAF-seq, researchers can perform single-molecule footprinting at near-nucleotide resolution, allowing for synchronous profiling of chromatin states and DNA sequences.
This technique reveals cooperative protein occupancy at individual regulatory elements and elucidates the functional impacts of somatic variants and rare chromatin epialleles. The single-cell DAF-seq approach generates comprehensive chromosome-length protein co-occupancy maps, capturing 99% of each cell’s mappable genome. Findings indicate that chromatin plasticity is extensive, with chromatin actuation diverging by as much as 61% between haplotypes within a single cell and 63% between different cells.
A.B. Stergachis, a key researcher in this study, highlighted that regulatory elements are preferentially co-actuated along the same chromatin fiber in a distance-dependent manner, mirroring the behavior of cohesin-mediated loops. This discovery underscores the intricate dynamics that govern gene regulation at a cellular level.
Implications for Gene Regulation Research
The implications of DAF-seq extend beyond basic research, as understanding chromatin fiber architectures can significantly impact the fields of genetics and epigenetics. By providing insights into how proteins interact with DNA, this technology could facilitate the identification of regulatory networks associated with various diseases, including cancer.
Researchers acknowledge the collaborative effort behind this advancement, noting contributions from various institutions and funding bodies, including the National Institutes of Health Common Fund and the Chan Zuckerberg Initiative. The study was co-authored by notable figures such as J.A. Stamatoyannopoulos and several other researchers who played pivotal roles in developing the DAF-seq workflows and analyzing the data.
The study, which was published in September 2023, represents a significant step forward in genomic research. As the scientific community continues to explore the complexities of gene regulation, the DAF-seq technique promises to shed light on the subtleties of chromatin behavior, paving the way for future discoveries in genetic engineering and therapy.
In summary, DAF-seq not only enhances our understanding of chromatin fiber architectures but also opens up new avenues for research and potential applications in biomedical science. The ability to map protein occupancy with single-cell precision marks a transformative leap in genetic research and holds promise for the future of personalized medicine.